mvNCProfile
Overview
mvNCProfile is a command line tool that compiles a network for use with the Intel® Movidius™ Neural Compute SDK (Intel® Movidius™ NCSDK), runs the network on a connected neural compute device, and outputs text and HTML profile reports.
The profiling data contains layer-by-layer statistics about the performance of the network. This is helpful in determining how much time is spent on each layer to narrow down potential changes to the network to improve the total inference time.
Syntax
Caffe
mvNCProfile network.prototxt [-w network.caffemodel] [-s max_number_of_shaves] [-in input_node_name] [-on output_node_name] [-is input_width input_height] [-ec]
TensorFlow*
mvNCProfile network.meta [-s max_number_of_shaves] [-in input_node_name] [-on output_node_name] [-is input_width input_height] [-ec]
| Argument | Description |
|---|---|
| Caffe: network.prototxt TensorFlow: network.meta network.pb |
Name of the network file (required). |
| [-w weights_file] | Specify the weights filename from training. For Caffe this is the .caffemodel file. If omitted, zero weights will be used. This option is not to be used for TensorFlow networks. |
| [-s max_number_of_shaves] | Specify the maximum number of SHAVEs to use for network layers (default: 1). The number of available SHAVEs depends on your neural compute device. The device runtime code may use fewer SHAVEs for some layers where measurements have typically shown no inference performance degradation (and consequently show a power benefit) from using fewer SHAVEs. |
| [-in input_node_name] | Specify an alternative start point for the network. By default the network’s start point is the input layer. This option enables partial network processing. When used together with the -on option, the user can isolate one or more layers in a network for analysis. This option is required for TensorFlow networks. You can use the name parameter (available for most layers) when creating your network and pass that name into this option. To add a named node that doesn’t change the network you can use the following: x = tensorflow.identity(prev_tensor, name='new_node') |
| [-on output_node_name] | Specify an alternative end point for the network. By default the network’s end point is the output layer. This option enables partial network processing. When used together with the -in option, the user can isolate one or more layers in a network for analysis. Be aware that the parser will stop at the first instance of this node name (e.g., a Relu following a Conv will not be processed if it shares the same name). This option is required for TensorFlow networks. You can use the name parameter (available for most layers) when creating your network and pass that name into this option. To add a named node that doesn’t change the network you can use the following: x = tensorflow.identity(prev_tensor, name='new_node') |
| [-is input_width input_height] | Specify input dimensions for networks that do not have dimension constraints on the input layer. This option assumes that the batch size is 1 and the number of channels is 3. |
| [-ec] | Skip certain compiler optimizations for concatenation; this may correct some issues with invalid results from concat layers or compile failures. |
| [–tf-ssd-config tensorflow_ssd_config_file] | Specify a TensorFlow SSD config file. This option is required to run TensorFlow SSD Mobilenet networks with the NCSDK. NCSDK TensorFlow SSD config file information. |
Examples
Caffe
mvNCProfile deploy.prototxt -w bvlc_googlenet.caffemodel -s 12 -in input -on prob -is 224 224
TensorFlow
mvNCProfile inception_v1.meta -s 12 -in input -on InceptionV1/Logits/Predictions/Reshape_1 -is 224 224
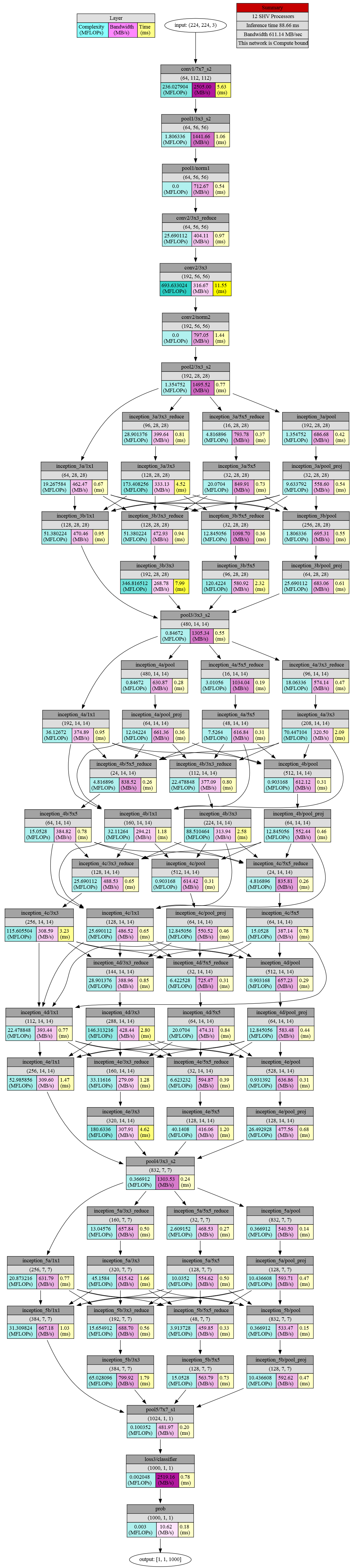
Example Profile Output for GoogLeNet
Text Format
Console output from the mvNCProfile tool is shown below.
Detailed Per Layer Profile
Layer Name MFLOPs Bandwidth MB/s time(ms)
========================================================================================
0 conv1/7x7_s2 236.028 2505.00 5.63
1 pool1/3x3_s2 1.806 1441.66 1.06
2 pool1/norm1 0.000 712.67 0.54
3 conv2/3x3_reduce 25.690 404.11 0.97
4 conv2/3x3 693.633 316.67 11.55
5 conv2/norm2 0.000 797.05 1.44
6 pool2/3x3_s2 1.355 1495.52 0.77
7 inception_3a/1x1 19.268 462.47 0.67
8 inception_3a/3x3_reduce 28.901 399.64 0.81
9 inception_3a/3x3 173.408 333.13 4.52
10 inception_3a/5x5_reduce 4.817 793.78 0.37
11 inception_3a/5x5 20.070 849.91 0.73
12 inception_3a/pool 1.355 686.68 0.42
13 inception_3a/pool_proj 9.634 558.60 0.54
14 inception_3b/1x1 51.380 470.46 0.95
15 inception_3b/3x3_reduce 51.380 472.93 0.94
16 inception_3b/3x3 346.817 268.78 7.99
17 inception_3b/5x5_reduce 12.845 1098.70 0.36
18 inception_3b/5x5 120.422 580.92 2.32
19 inception_3b/pool 1.806 695.31 0.55
20 inception_3b/pool_proj 25.690 683.06 0.61
21 pool3/3x3_s2 0.847 1305.34 0.55
22 inception_4a/1x1 36.127 374.89 0.95
23 inception_4a/3x3_reduce 18.063 574.14 0.47
24 inception_4a/3x3 70.447 320.50 2.09
25 inception_4a/5x5_reduce 3.011 1034.04 0.19
26 inception_4a/5x5 7.526 616.84 0.31
27 inception_4a/pool 0.847 630.87 0.28
28 inception_4a/pool_proj 12.042 661.36 0.36
29 inception_4b/1x1 32.113 294.21 1.18
30 inception_4b/3x3_reduce 22.479 377.09 0.80
31 inception_4b/3x3 88.510 313.94 2.58
32 inception_4b/5x5_reduce 4.817 838.52 0.26
33 inception_4b/5x5 15.053 384.82 0.78
34 inception_4b/pool 0.903 612.12 0.31
35 inception_4b/pool_proj 12.845 552.44 0.46
36 inception_4c/1x1 25.690 486.52 0.65
37 inception_4c/3x3_reduce 25.690 488.53 0.65
38 inception_4c/3x3 115.606 308.59 3.23
39 inception_4c/5x5_reduce 4.817 835.81 0.26
40 inception_4c/5x5 15.053 387.14 0.78
41 inception_4c/pool 0.903 614.42 0.31
42 inception_4c/pool_proj 12.845 550.52 0.46
43 inception_4d/1x1 22.479 393.44 0.77
44 inception_4d/3x3_reduce 28.901 388.96 0.85
45 inception_4d/3x3 146.313 428.44 2.80
46 inception_4d/5x5_reduce 6.423 725.47 0.31
47 inception_4d/5x5 20.070 474.31 0.84
48 inception_4d/pool 0.903 657.23 0.29
49 inception_4d/pool_proj 12.845 583.48 0.44
50 inception_4e/1x1 52.986 309.60 1.47
51 inception_4e/3x3_reduce 33.116 279.09 1.28
52 inception_4e/3x3 180.634 307.91 4.62
53 inception_4e/5x5_reduce 6.623 594.87 0.39
54 inception_4e/5x5 40.141 416.06 1.20
55 inception_4e/pool 0.931 636.86 0.31
56 inception_4e/pool_proj 26.493 477.56 0.68
57 pool4/3x3_s2 0.367 1303.53 0.24
58 inception_5a/1x1 20.873 631.79 0.77
59 inception_5a/3x3_reduce 13.046 657.84 0.50
60 inception_5a/3x3 45.158 615.42 1.66
61 inception_5a/5x5_reduce 2.609 468.53 0.27
62 inception_5a/5x5 10.035 554.62 0.50
63 inception_5a/pool 0.367 540.50 0.14
64 inception_5a/pool_proj 10.437 593.71 0.47
65 inception_5b/1x1 31.310 667.18 1.03
66 inception_5b/3x3_reduce 15.655 688.70 0.56
67 inception_5b/3x3 65.028 799.92 1.79
68 inception_5b/5x5_reduce 3.914 459.85 0.33
69 inception_5b/5x5 15.053 563.79 0.73
70 inception_5b/pool 0.367 533.47 0.15
71 inception_5b/pool_proj 10.437 592.62 0.47
72 pool5/7x7_s1 0.100 481.97 0.20
73 loss3/classifier 0.002 2519.16 0.78
74 prob 0.003 10.62 0.18
----------------------------------------------------------------------------------------
Total inference time 88.66
----------------------------------------------------------------------------------------
Graphical Format
The graphical representation of the profile information (saved as output_report.html and output.gv.svg files) is shown below.